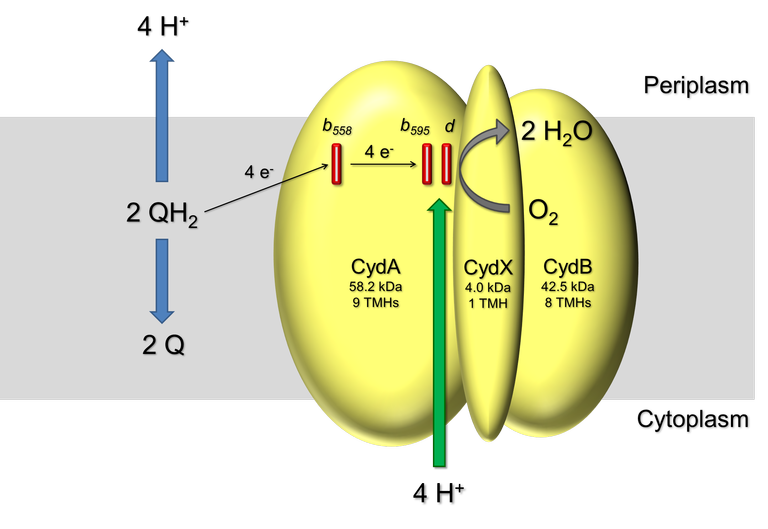
Cytochrome bd ubiquinol oxidases are terminal respiratory oxidases of many prokaryotes, including several pathogens. The enzyme complex couples (ubi)quinol oxidation and release of protons to the periplasmic side with proton uptake from the cytoplasmic side to reduce dioxygen to water. In doing so, bd oxidases contribute to the generation of the protonmotive force by vectorial charge transfer. Consistently, bd oxidase is made up of two major subunits CydA and B that share the same fold of two four-helix bundles and an additional cytoplasmic helix. CydA harbours the cofactors hemes b558, b595, and d. Heme b558accepts electrons from (ubi)quinol and transfers them via heme b595 to the active site composed of heme d, where dioxygen is reduced to water. In addition, CydA features a globular domain located on the periplasmic side. This region, termed Q-loop, is expected to be involved in binding and oxidation of the substrate (ubi)quinol. In dependence of its length, the family of bdoxidases is classified into the S (short)- and L (large)-subfamilies.
Beyond, it was found that bd oxidases contain a third subunit, called CydX in Escherichia coli and CydS in Geobacillus thermodenitrificans. This small subunit is encoded in the cydoperon and it probably stabilizes the active center. Surprisingly, E. coli bd oxidase contains a forth subunit called either CydY or CydH that is not encoded in the cyd operon but derives from the orphan gene ynhF. Surprisingly, the G. thermodenitrificansenzyme lacks a corresponding subunit. Remarkably, this subunit blocks the dioxygen entry site found in both enzymes. However, the E. coli enzyme contains an additional, unique substrate channel leading from CydB to heme d. In accordance with the different positions of the dioxygen binding sites, the positions of heme b595and d are interchanged in the E. coli enzyme with respect to the G. thermodenitrificans oxidase. This implies that the homologous enzymes may have different physiological functions, hence working either as a true oxidase (in E. coli) or as a bacterial defense factor (in G. denitrificans).
References
Hoeser, J., Hong, S., Gehmann, G., Gennis, R.B., Friedrich, T. (2014) Subunit CydX of Escherichia colicytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett. 588, 1537-41.
Theßeling, A., Rasmussen, T., Burschel, S., Wohlwend, D., Kägi, J., Müller, R., Böttcher, B., and Friedrich, T. (2019) Homologous bd oxidases share the same architecture but differ in mechanism. Nat. Comm., 10: 5138. DOI: 10.1038/s41467-019-13122-4.
Grauel, A., Kägi, J., Rasmussen, T., Makarchuk, I., Oppermann, S., Moumbock, A.F.A., Wohlwend, D., Müller, R., Melin, F., Günther, S., Hellwig, P., Böttcher, B. and Friedrich, T. (2021) Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D. Nat. Commun., 12:6498. doi.org/10.1038/s41467-021-26835-2.
Friedrich, T., Wohlwend, D. and Borisov, V.B. (2022) Recent Advances in Structural Studies of Cytochrome bd and Its Potential Application as a Drug Target. Int. J. Mol. Sci. 23, 3166. doi.org/10.3390/ijms23063166


